Deciphering molecular networks in health and disease by leveraging AI-based protein structure prediction and multi-omics integration
Welcome to the website of the research group:
Molecular Biology of Signalling in Inflammatory Processes
Molecular Pharmacology, Aachen
Our goal is to unravel the molecular mechanisms that regulate cell signalling and to understand how signalling controls biological functions in health and disease.
To achieve this, we have developed a computational pipeline MaPIN that integrates AI-based protein structure prediction (AlphaFold) with multi-omics data to unravel molecular networks and pathways at a functional level.
NEWS
NEWS
We are happy to share that our research on the proteolytic release of angiotensin-converting enzyme (ACE) from endothelial cells has been published in the FASEB Journal.
10 October 2024
Webers M., Y. Yu, J. Eyll, J. Vanderliek-Kox, K. Schun, A. Michely, T. Schumertl, C. Garbers, J. Dietrich, D. D. Jonigk, I. Krüger, M. P. Kühnel, C. Martin, A. Ludwig, Stefan Düsterhöft (2024). “The metalloproteinase ADAM10 sheds angiotensin-converting enzyme (ACE) from the pulmonary endothelium as a soluble, functionally active convertase." FASEBJ doi.org/10.1096/fj.202402069R
7 October 2024
We are very happy that our grant proposal “MaPION” (Mapping Protein Interactions in Oncologic Networks) in the call DataXperiment was selected to be funded by the BMBF - Nationale Dekade gegen Krebs.
MaPIN
Mapping of Protein Interaction Networks
Recent advances in technology have greatly improved our ability to study genes and proteins in cells, but understanding how these discoveries affect biological processes remains a challenge. Many proteins function in intricate clusters and networks where protein functions and their signals cross and modulate each other, making it essential to unravel these interactions to better understand complex physiological and pathological conditions.
AI tools such as AlphaFold, which predicts protein structures, have revolutionised our understanding of protein function. Building on this, we have developed MaPIN (Mapping of Protein Interaction Networks), a computational tool designed to analyse protein interactions in a variety of biological contexts. By integrating data from protein studies, gene expression and molecular mutations, MaPIN helps to identify critical protein networks and interaction sites.
What sets MaPIN apart is its ability to look beyond established pathways and uncover novel interactions within these complex networks. This approach provides deeper insights into the underlying mechanisms of different physiological processes and disease states, potentially revealing new therapeutic targets.
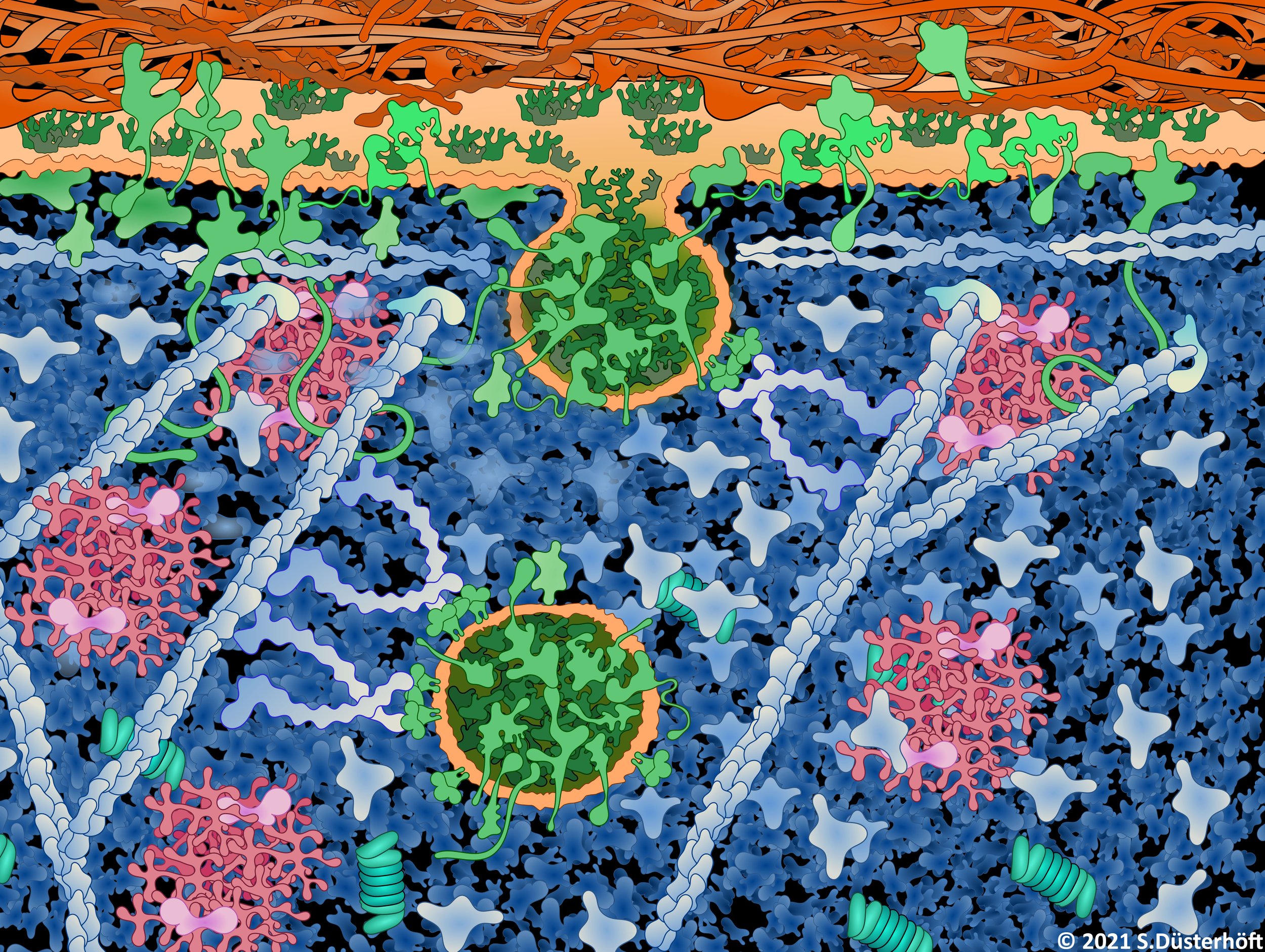
Cluster Biology of Inflammation
Like the iRhom-ADAM17 complex, proteins are often part of larger functional complexes. When analysing these complexes, one has to take into account that these complexes are not static, but rather change dynamically depending on the cellular milieu and functional requirements. Therefore, we tend to consider the target proteins in our studies as parts of larger molecular clusters.
Do It Yourself Labware
3D-printing has enabled us to produce low-cost laboratory items.
We are happy to share our designs!
If you are interested in the possibilities of 3D-printed labware, simply contact us.
Projects

















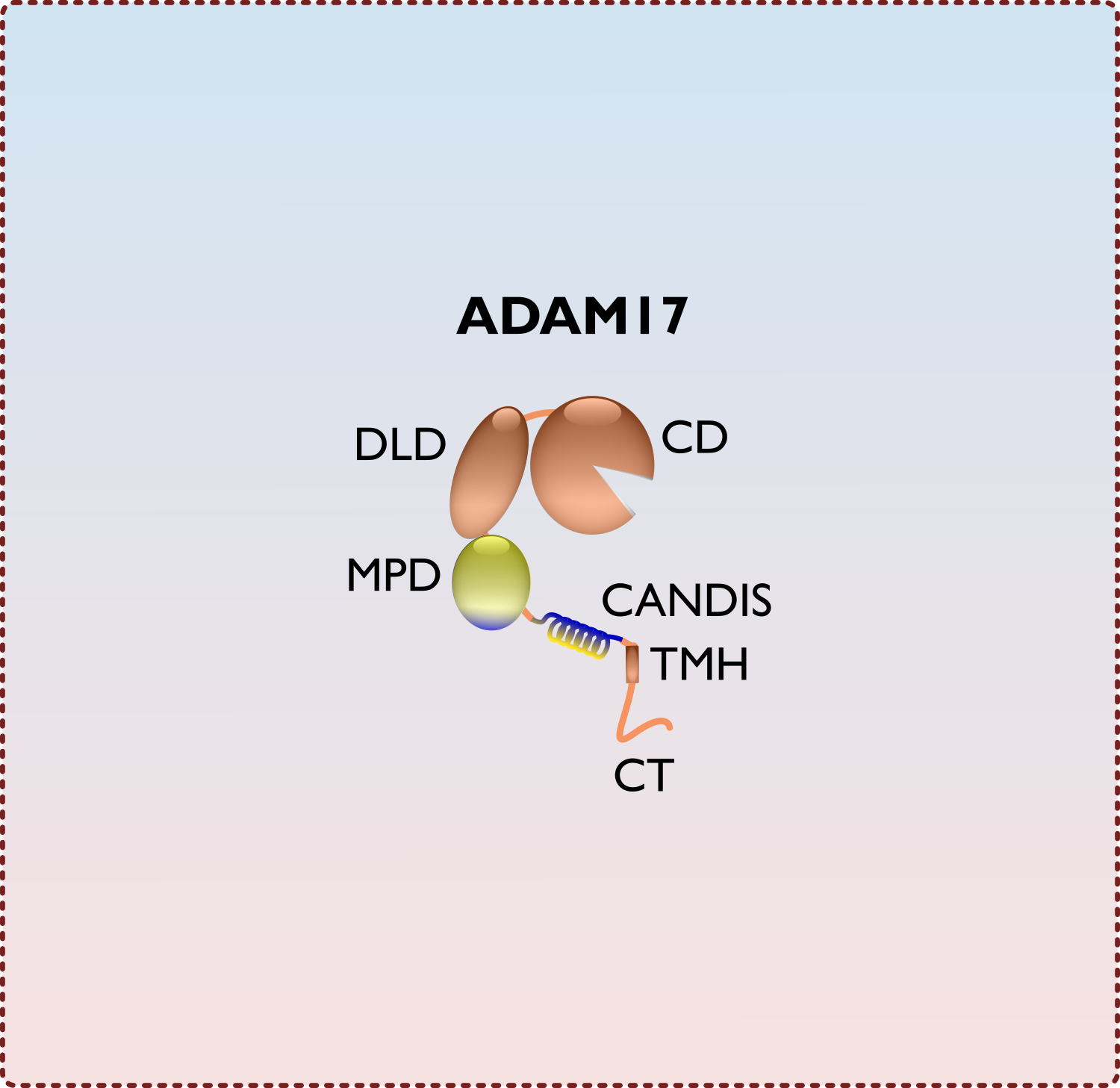

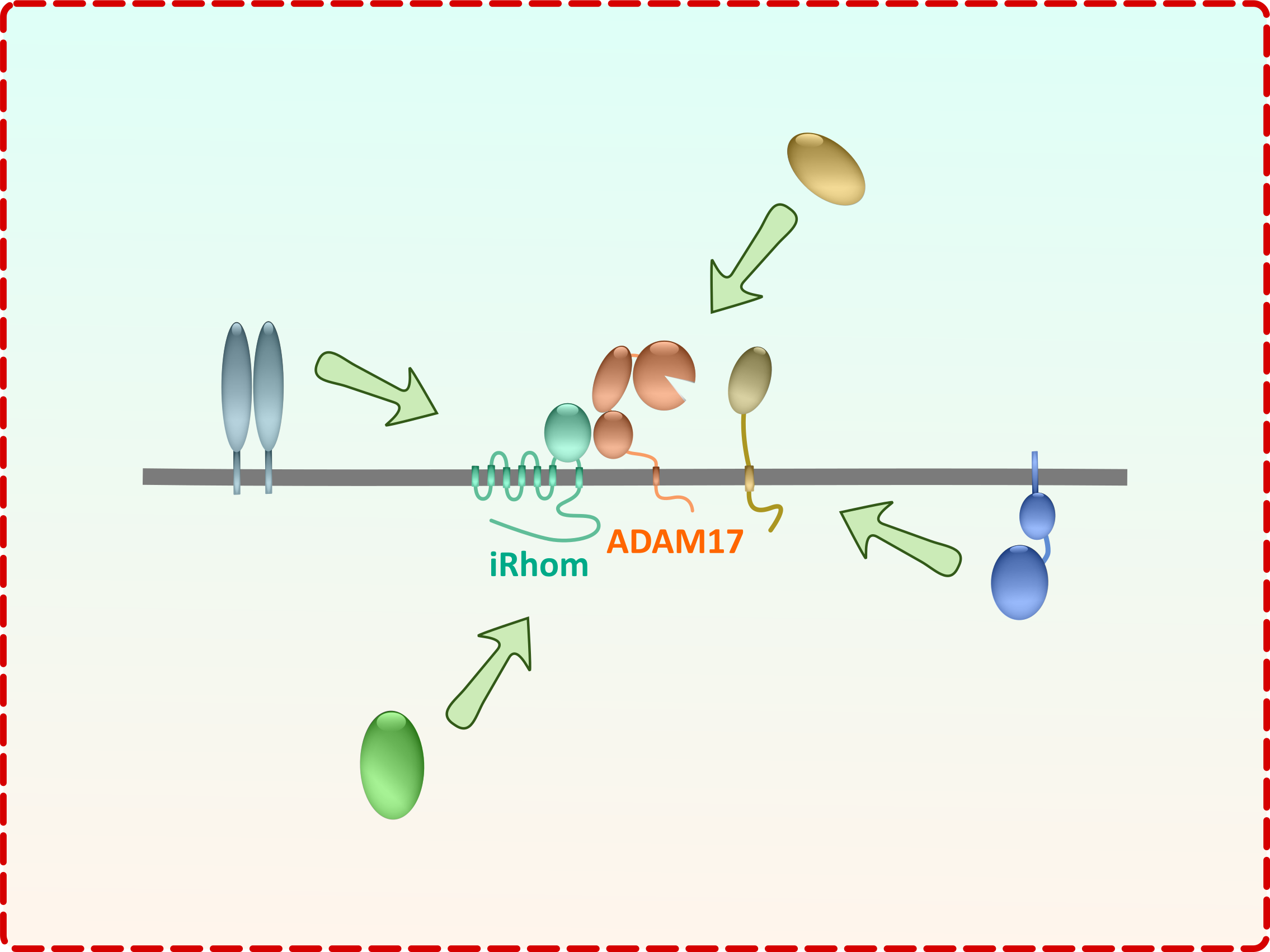

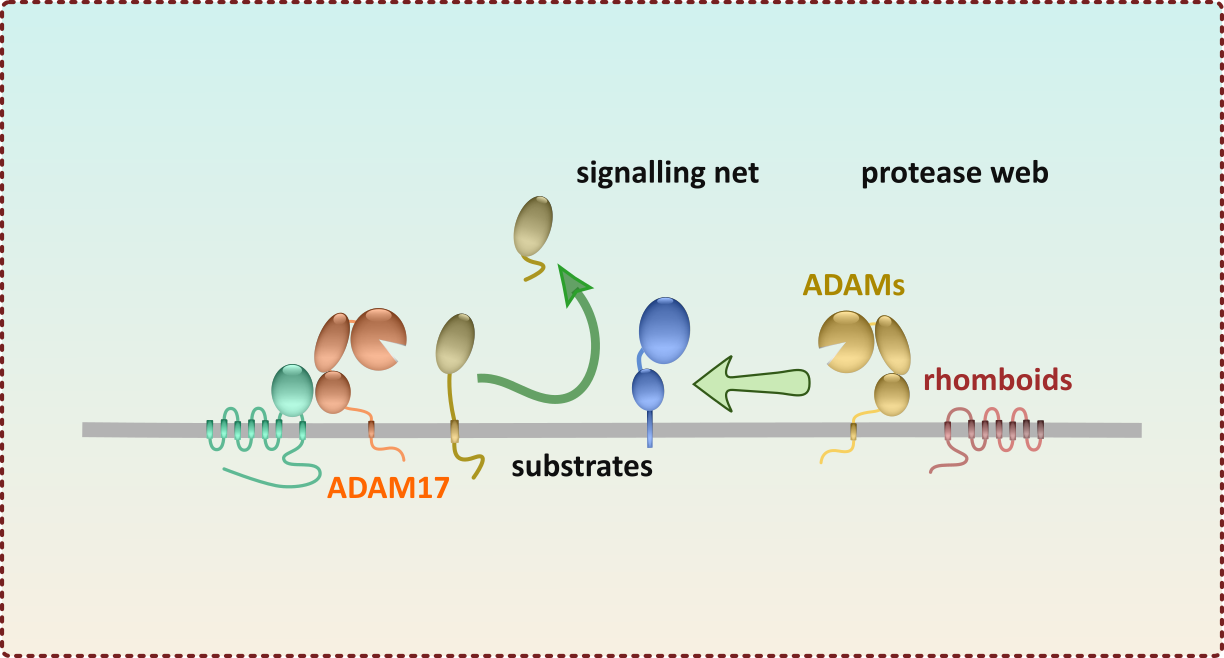
Life-cycle of the iRhom-ADAM17 shedding complex